Visualizing vibronic transitions in a diatomic molecule
Posted on 07 October 2020
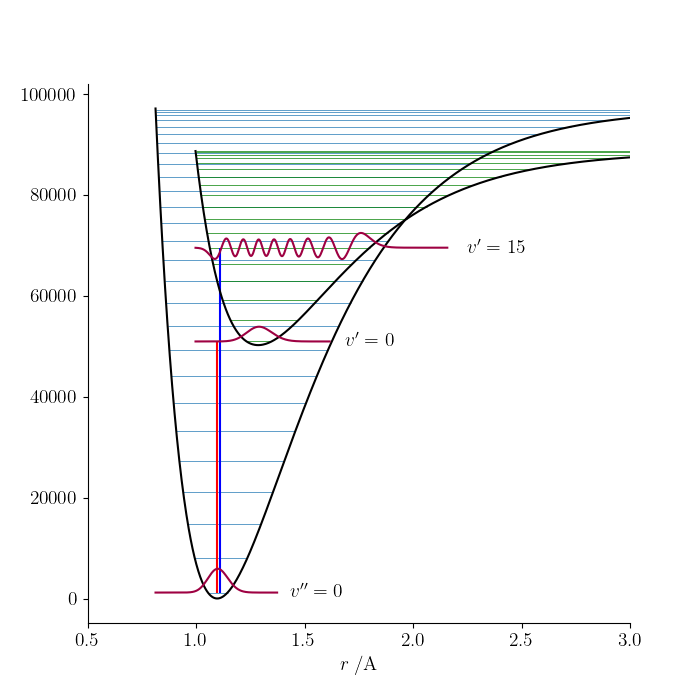
The Morse oscillator code introduced in a previous blog post can be used to visualize the vibronic transitions in a diatomic molecule by creating two Morse objects (one for each electronic state) and plotting their potential energy curves and energy levels on the same Matplotlib Axes object.
import numpy as np
from matplotlib import rc
import matplotlib.pyplot as plt
from scipy.constants import h, c
from morse import Morse, FAC
rc('font', **{'family': 'serif', 'serif': ['Computer Modern'], 'size': 14})
rc('text', usetex=True)
# Figure resolution and size in pixels.
DPI = 100
w, h = 700, 525
# The colour to plot wavefunctions in.
COLOUR1 = (0.6196, 0.0039, 0.2588, 1.0)
# Atom masses and ground state parameters for (14N)2.
mA = mB = 14.003074004
X_re = 1.09768e-10
X_Te = 0
X_we, X_wexe = 2358.57, 14.324
X = Morse(mA, mB, X_we, X_wexe, X_re, X_Te)
X.make_rgrid(rmax=3.e-10)
X.V = X.Vmorse(X.r)
# First excited electronic state parameters for (14N)2.
A_re = 1.2866e-10
A_Te = 50203.6
A_we, A_wexe = 1460.64, 13.87
A = Morse(mA, mB, A_we, A_wexe, A_re, A_Te)
fig, ax = plt.subplots(figsize=(w/DPI, w/DPI), dpi=DPI)
# Plot the ground state potential energy curve and vibrational energy levels.
X.plot_V(ax, color='k')
X.draw_Elines(range(0, X.vmax, 3), ax, lw=0.5)
# Draw and label the ground state vibrational wavefunction.
X.plot_psi([0], ax, scaling=4, color=COLOUR1)
X.label_levels([0], ax, [r"$v''=0$"])
# Plot the excited state potential energy curve and vibrational energy levels.
A.plot_V(ax, color='k')
A.draw_Elines(range(0, A.vmax, 3), ax, lw=0.5, color='g')
# Draw and label the excited state vibrational wavefunctions for v=0 and v=15.
A.plot_psi([0, 15], ax, scaling=4, color=COLOUR1)
A.label_levels([0, 15], ax, [r"$v'=0$", r"$v'=15$"])
# Some figure labelling and tidying.
ax.set_xlabel(r'$r \;/ \mathrm{\AA}$')
ax.spines['top'].set_visible(False)
ax.spines['right'].set_visible(False)
# Indicate a couple of transitions.
Epp = X.Emorse(0, 'cm-1', include_Te=True)
Ep = A.Emorse(0, 'cm-1', include_Te=True)
ax.vlines(X.re*1.e10, Epp, Ep, color='r')
Ep = A.Emorse(15, 'cm-1', include_Te=True)
ax.vlines(X.re*1.e10*1.01, Epp, Ep, color='b')
ax.set_xlim(0.5, 3)
plt.savefig('morse-vibronic.png', dpi=DPI)
plt.show()
There are a couple of changes to the original code, which are included in the updated version of morse.py here:
import numpy as np
from scipy.constants import h, hbar, c, u
from scipy.special import factorial
from scipy.special import genlaguerre, gamma
# Factor for conversion from cm-1 to J
FAC = 100 * h * c
class Morse:
"""A class representing the Morse oscillator model of a diatomic."""
def __init__(self, mA, mB, we, wexe, re, Te):
"""Initialize the Morse model for a diatomic molecule.
mA, mB are the atom masses (atomic mass units).
we, wexe are the Morse parameters (cm-1).
re is the equilibrium bond length (m).
Te is the electronic energy (minimum of the potential well; origin
of the vibrational state energies).
"""
self.mA, self.mB = mA, mB
self.mu = mA*mB/(mA+mB) * u
self.we, self.wexe = we, wexe
self.re = re
self.Te = Te
self.De = we**2 / 4 / wexe * FAC
self.ke = (2 * np.pi * c * 100 * we)**2 * self.mu
# Morse parameters, a and lambda.
self.a = self.calc_a()
self.lam = np.sqrt(2 * self.mu * self.De) / self.a / hbar
# Maximum vibrational quantum number.
self.vmax = int(np.floor(self.lam - 0.5))
self.make_rgrid()
self.V = self.Vmorse(self.r)
def make_rgrid(self, n=1000, rmin=None, rmax=None, retstep=False):
"""Make a suitable grid of internuclear separations."""
self.rmin, self.rmax = rmin, rmax
if rmin is None:
# minimum r where V(r)=De on repulsive edge
self.rmin = self.re - np.log(2) / self.a
if rmax is None:
# maximum r where V(r)=f.De
f = 0.999
self.rmax = self.re - np.log(1-f)/self.a
self.r, self.dr = np.linspace(self.rmin, self.rmax, n,
retstep=True)
if retstep:
return self.r, self.dr
return self.r
def calc_a(self):
"""Calculate the Morse parameter, a.
Returns the Morse parameter, a, from the equilibrium
vibrational wavenumber, we in cm-1, and the dissociation
energy, De in J.
"""
return (self.we * np.sqrt(2 * self.mu/self.De) * np.pi *
c * 100)
def Vmorse(self, r):
"""Calculate the Morse potential, V(r).
Returns the Morse potential at r (in m) for parameters De
(in J), a (in m-1) and re (in m).
"""
return self.De * (1 - np.exp(-self.a*(r - self.re)))**2
def Emorse(self, v, units='J', include_Te=False):
"""Calculate the energy of a Morse oscillator in state v.
Returns the energy of a Morse oscillator parameterized by
equilibrium vibrational frequency we and anharmonicity
constant, wexe (both in cm-1).
"""
vphalf = v + 0.5
E = self.we * vphalf - self.wexe * vphalf**2
if include_Te:
E += self.Te
if units == 'cm-1':
return E
elif units == 'J':
return E * FAC
raise ValueError('unrecognised units "{}" in Emorse'.format(units))
def calc_turning_pts(self, E):
"""Calculate the classical turning points at energy E.
Returns rm and rp, the classical turning points of the Morse
oscillator at energy E (provided in J). rm < rp.
"""
b = np.sqrt(E / self.De)
return (self.re - np.log(1+b) / self.a,
self.re - np.log(1-b) / self.a)
def calc_psi(self, v, r=None, normed=True, psi_max=1):
"""Calculates the Morse oscillator wavefunction, psi_v.
Returns the Morse oscillator wavefunction at vibrational
quantum number v. The returned function is "normalized" to
give peak value psi_max.
"""
if r is None:
r = self.r
z = 2 * self.lam * np.exp(-self.a*(r - self.re))
alpha = 2*(self.lam - v) - 1
psi = (z**(self.lam-v-0.5) * np.exp(-z/2) *
genlaguerre(v, alpha)(z))
psi *= psi_max / np.max(psi)
return psi
def calc_psi_z(self, v, z):
alpha = 2*(self.lam - v) - 1
psi = (z**(self.lam-v-0.5) * np.exp(-z/2) *
genlaguerre(v, alpha)(z))
Nv = np.sqrt(factorial(v) * (2*self.lam - 2*v - 1) /
gamma(2*self.lam - v))
return Nv * psi
def plot_V(self, ax, **kwargs):
"""Plot the Morse potential on Axes ax."""
ax.plot(self.r*1.e10, self.V / FAC + self.Te, **kwargs)
def get_vmax(self):
"""Return the maximum vibrational quantum number."""
return int(self.we / 2 / self.wexe - 0.5)
def draw_Elines(self, vlist, ax, **kwargs):
"""Draw lines on Axes ax representing the energy level(s) in vlist."""
if isinstance(vlist, int):
vlist = [vlist]
for v in vlist:
E = self.Emorse(v)
rm, rp = self.calc_turning_pts(E)
ax.hlines(E / FAC + self.Te, rm*1.e10, rp*1e10, **kwargs)
def label_levels(self, vlist, ax, labels=None):
if isinstance(vlist, int):
vlist = [vlist]
if labels is None:
labels = [r'$v={}$'.format(v) for v in vlist]
for i, v in enumerate(vlist):
E = self.Emorse(v)
rm, rp = self.calc_turning_pts(E)
ax.text(s=labels[i], x=rp*1e10*1.25,
y=E / FAC + self.Te, va='center')
def plot_psi(self, vlist, ax, r_plot=None, scaling=1, **kwargs):
"""Plot the Morse wavefunction(s) in vlist on Axes ax."""
if isinstance(vlist, int):
vlist = [vlist]
for v in vlist:
E = self.Emorse(v)
if r_plot is None:
rm, rp = self.calc_turning_pts(E)
x = self.r[self.r<rp*1.2]
else:
x = r_plot
psi = self.calc_psi(v, r=x, psi_max=self.we/2)
psi_plot = psi*scaling + self.Emorse(v)/FAC + self.Te
ax.plot(x*1.e10, psi_plot, **kwargs)