Posted by: christian on 19 Dec 2018
The scipy.optimize.curve_fit routine can be used to fit two-dimensional data, but the fitted data (the ydata argument) must be repacked as a one-dimensional array first. The independent variable (the xdata argument) must then be an array of shape (2,M) where M is the total number of data points.
For a two-dimensional array of data, Z, calculated on a mesh grid (X, Y), this can be achieved efficiently using the ravel method:
xdata = np.vstack((X.ravel(), Y.ravel()))
ydata = Z.ravel()
The following code demonstrates this approach for some synthetic data set created as a sum of four Gaussian functions with some noise added:
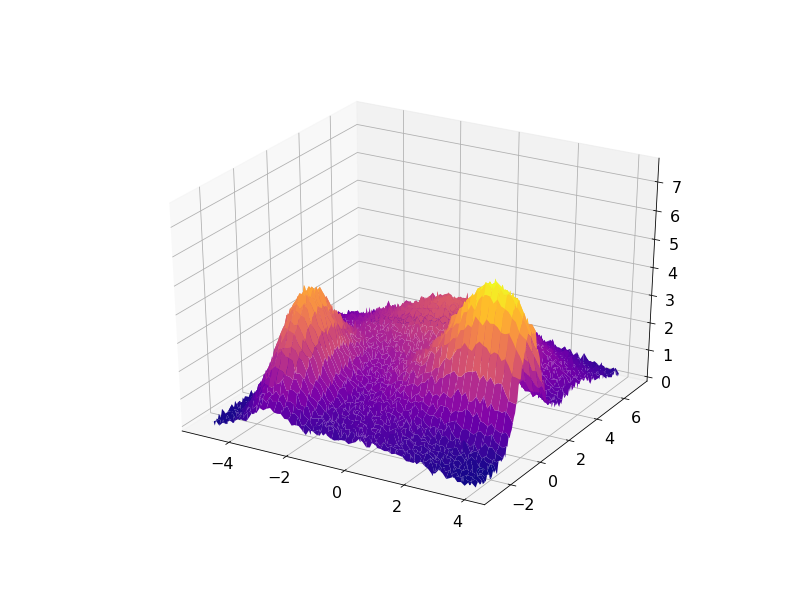
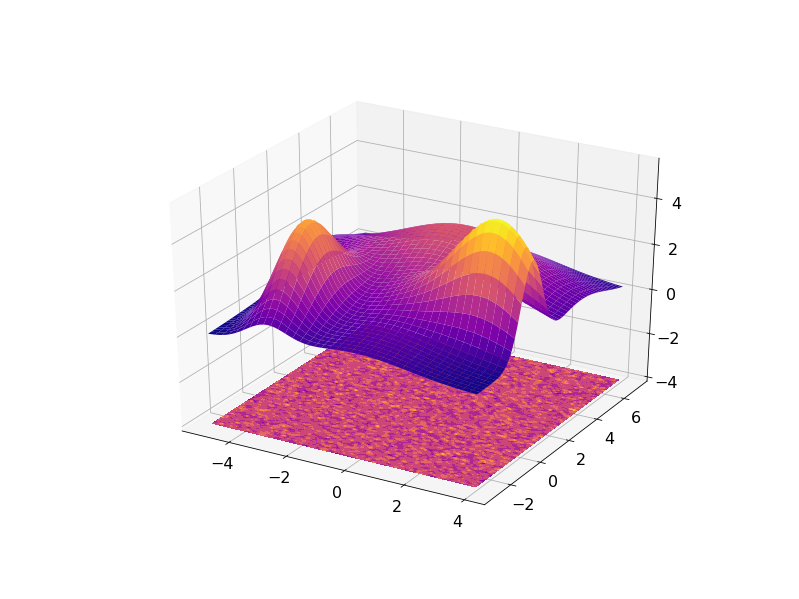
The result can be visualized in 3D with the residuals plotted on a plane under the fitted data:
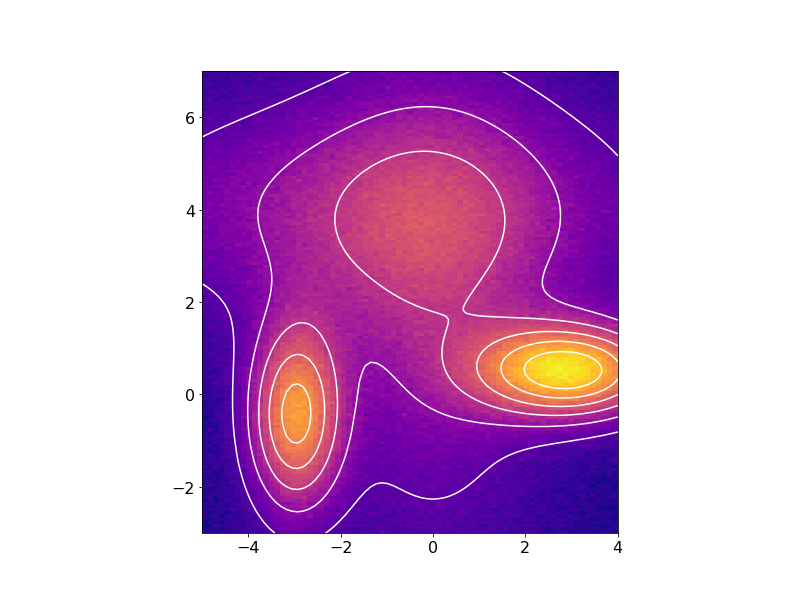
or in 2D with the fitted data contours superimposed on the noisy data:
import numpy as np
from scipy.optimize import curve_fit
import matplotlib.pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
# The two-dimensional domain of the fit.
xmin, xmax, nx = -5, 4, 75
ymin, ymax, ny = -3, 7, 150
x, y = np.linspace(xmin, xmax, nx), np.linspace(ymin, ymax, ny)
X, Y = np.meshgrid(x, y)
# Our function to fit is going to be a sum of two-dimensional Gaussians
def gaussian(x, y, x0, y0, xalpha, yalpha, A):
return A * np.exp( -((x-x0)/xalpha)**2 -((y-y0)/yalpha)**2)
# A list of the Gaussian parameters: x0, y0, xalpha, yalpha, A
gprms = [(0, 2, 2.5, 5.4, 1.5),
(-1, 4, 6, 2.5, 1.8),
(-3, -0.5, 1, 2, 4),
(3, 0.5, 2, 1, 5)
]
# Standard deviation of normally-distributed noise to add in generating
# our test function to fit.
noise_sigma = 0.1
# The function to be fit is Z.
Z = np.zeros(X.shape)
for p in gprms:
Z += gaussian(X, Y, *p)
Z += noise_sigma * np.random.randn(*Z.shape)
# Plot the 3D figure of the fitted function and the residuals.
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot_surface(X, Y, Z, cmap='plasma')
ax.set_zlim(0,np.max(Z)+2)
plt.show()
# This is the callable that is passed to curve_fit. M is a (2,N) array
# where N is the total number of data points in Z, which will be ravelled
# to one dimension.
def _gaussian(M, *args):
x, y = M
arr = np.zeros(x.shape)
for i in range(len(args)//5):
arr += gaussian(x, y, *args[i*5:i*5+5])
return arr
# Initial guesses to the fit parameters.
guess_prms = [(0, 0, 1, 1, 2),
(-1.5, 5, 5, 1, 3),
(-4, -1, 1.5, 1.5, 6),
(4, 1, 1.5, 1.5, 6.5)
]
# Flatten the initial guess parameter list.
p0 = [p for prms in guess_prms for p in prms]
# We need to ravel the meshgrids of X, Y points to a pair of 1-D arrays.
xdata = np.vstack((X.ravel(), Y.ravel()))
# Do the fit, using our custom _gaussian function which understands our
# flattened (ravelled) ordering of the data points.
popt, pcov = curve_fit(_gaussian, xdata, Z.ravel(), p0)
fit = np.zeros(Z.shape)
for i in range(len(popt)//5):
fit += gaussian(X, Y, *popt[i*5:i*5+5])
print('Fitted parameters:')
print(popt)
rms = np.sqrt(np.mean((Z - fit)**2))
print('RMS residual =', rms)
# Plot the 3D figure of the fitted function and the residuals.
fig = plt.figure()
ax = fig.gca(projection='3d')
ax.plot_surface(X, Y, fit, cmap='plasma')
cset = ax.contourf(X, Y, Z-fit, zdir='z', offset=-4, cmap='plasma')
ax.set_zlim(-4,np.max(fit))
plt.show()
# Plot the test data as a 2D image and the fit as overlaid contours.
fig = plt.figure()
ax = fig.add_subplot(111)
ax.imshow(Z, origin='bottom', cmap='plasma',
extent=(x.min(), x.max(), y.min(), y.max()))
ax.contour(X, Y, fit, colors='w')
plt.show()
Comments
Comments are pre-moderated. Please be patient and your comment will appear soon.
Dominik Stańczak 5 years, 5 months ago
That's a pretty darn clever solution! I'm absolutely stealing this code. :)
Link | ReplyGonzalo Velarde 4 years, 4 months ago
Thank you for that excelent approach!
Link | Replywhat if I have "nan" in my Z grid?
Is convinient to replace them with zeros?
Z[numpy.isnan(Z)]=0
or is it better to convert ndarrays into linear arrays
taking out zero values?
x=[]
y=[]
z=[]
for j in range(1,len(y)):
for i in range(1,len(x)):
if z_with_zeros[i][j]==0:
pass
else:
x.append(x[i][j])
y.append(y[i][j])
z.append(z[i][j])
thank you in advance
Martin Rommel 1 year, 3 months ago
curve_fit does not rely on a regular data grid. You just give it a list of points. Therefore you can simply remove the nan point from xdata and Z.ravel()
Link | Replychristian 4 years, 4 months ago
You probably don't want to set them to zero, since you're fitted surface (curve) will try to go through zero there as a value of the input data and bias the fit. Have you tried interpolating the missing values before the fit?
Link | ReplyPhilip Phishgills 3 years, 8 months ago
What if you don't know what function you want to fit? Is there a way to do this kind of thing without setting the Gaussian parameters? (say I know it's a sum of 10 Gaussians, but I'm not sure about their parameters)
Link | Replychristian 3 years, 8 months ago
If you know the function you want to fit but not the parameters, and the function is non-linear in those parameters, then you likely need some initial guesses to the parameters to set the fitting routine off. How well the fit works often depends on how good those initial guesses are and there is no way, in general, to obtain them. A good start is to plot your function and look for inspiration there (e.g. find the Gaussian centres). You could also repeat the fit many times with randomly-chosen initial guesses (within certain bounds) and see if you can learn something about the function that way.
Link | ReplyThere are some more comments about this issue in this question: https://stats.stackexchange.com/questions/62995/how-to-choose-initial-values-for-nonlinear-least-squares-fit
Rafael 2 years, 6 months ago
That is excellent. How would this look like if the function was a 2D polynomial?
Link | ReplyI'm trying to apply this using numpy's poly2d
the function itself is
polyval2d(X,Y,C)
where C is a (n,m) coefficient matrix.
christian 2 years, 5 months ago
Thank you.
Link | ReplyFitting to a polynomial is, in principle, a linear least squares problem – you could look at https://scipython.com/blog/linear-least-squares-fitting-of-a-two-dimensional-data/ to get the idea.
L Gee 2 years, 2 months ago
Really good solution, absolutely using this. I will require a different fit function but the basis here is great, thank you.
Link | ReplyNew Comment