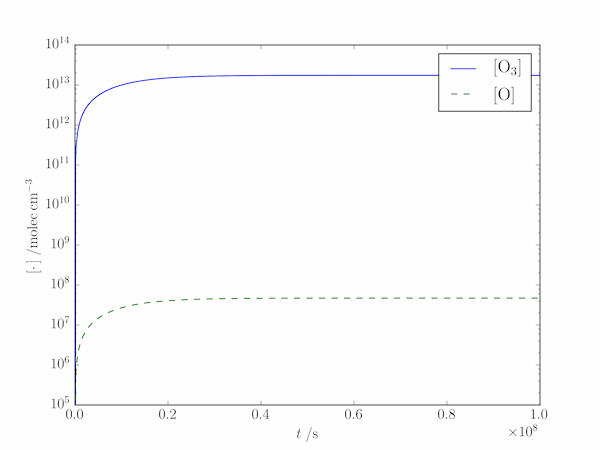
Solution P8.2.6
The code below uses scipy.integrate.odeint to solve the three coupled differential equations describing the concentrations of $\mathrm{O_2}$, $\mathrm{O}$ and $\mathrm{O_3}$ with time.
import numpy as np
from scipy.integrate import odeint
import pylab
# Reaction rate constants
k1 = 3.e-12 # O2 + hv -> 2O (s-1)
k2 = 1.2e-33 # O2 + O + M -> O3 + M (cm6.molec-2.s-1)
k3 = 5.5e-4 # O3 + hv' -> O + O2 (s-1)
k4 = 6.9e-16 # O + O3 -> 2O2 (cm3.molec-1.s-1)
def d(c, t, M):
""" Return the d[X]/dt for each species. """
O2, O, O3 = c
dO2dt = -k1*O2 - k2*M*O*O2 + k3*O3 + 2*k4*O*O3
dOdt = 2*k1*O2 - k2*M*O*O2 + k3*O3 - k4*O*O3
dO3dt = k2*M*O*O2 - k3*O3 - k4*O*O3
return dO2dt, dOdt, dO3dt
# Total molecule concentration, M, and O2 concentration, cO2
M = 9.e17
cO2 = 0.21*M
# Initial conditions for [O2], [O], [O3]
c0 = [cO2, 0, 0]
# Integrate the differential equations over a suitable time grid (s)
t = np.linspace(0, 1.e8, 1000)
c = odeint(d, c0, t, args=(M,))
# Steady-state approximation solution for comparison
cO3ss = np.sqrt(k1 * k2 / k3 / k4 * M) * cO2
cOss = k3 * cO3ss / k2 / cO2 / M
print('Numerical values:\n[O] = {:g} molec/cm3, [O3] = {:g} molec/cm3'
.format(*c[-1][1:]))
print('Steady-state values:\n[O]ss = {:g} molec/cm3, [O3]ss = {:g} molec/cm3'
.format(cOss, cO3ss))
# Plot the evolution of [O3] and [O] with time
pylab.plot(t,c[:,2], c='k', label=r'$\mathrm{[O_3]}$')
pylab.plot(t,c[:,1], c='k', ls='--', label=r'$\mathrm{[O]}$')
pylab.yscale('log')
pylab.xlabel(r'$t\;/\mathrm{s}$')
pylab.ylabel(r'$[\cdot\,]\;/\mathrm{molec\,cm^{-3}}$')
pylab.legend()
pylab.show()
The output shows that the steady-state approximation works well at this altitude:
Numerical values:
[O] = 4.70561e+07 molec/cm3, [O3] = 1.74604e+13 molec/cm3
Steady-state values:
[O]ss = 4.7055e+07 molec/cm3, [O3]ss = 1.74634e+13 molec/cm3