Power Spectra for "Blue" and Uniform Noise
Posted on 24 April 2017
The previous post provided some code for generating Poisson disc noise (in which no point the sample is closer than some fixed distance from any other). Here is a short program to calculate the power spectrum of this noise and compare it with the spectrum for the same number of points drawn from a uniform distribution.
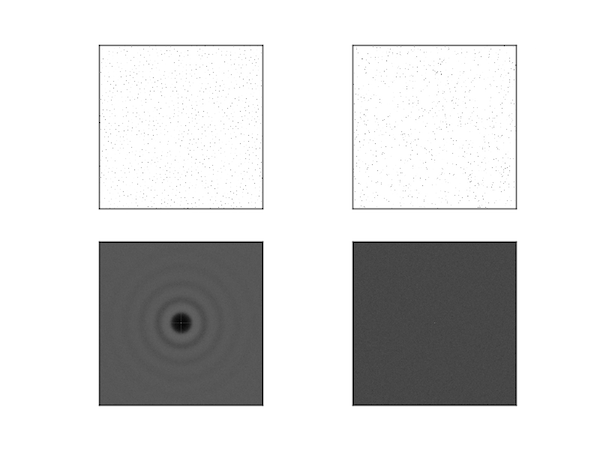
Top: Poisson disc noise (left) and uniform noise (right) and their power spectra (bottom). As expected, the spectrum of the uniform noise is featureless, whereas the spectrum for Poisson disc noise has concentric circles around a blank disc.
The code below uses a version of the Poisson Disc algorithm that I have cast into object-oriented form (also given below).
This code is also available on my github page.
import numpy as np
import matplotlib.pyplot as plt
from poisson import PoissonDisc
class UniformNoise():
"""A class for generating uniformly distributed, 2D noise."""
def __init__(self, width=50, height=50, n=None):
"""Initialise the size of the domain and number of points to sample."""
self.width, self.height = width, height
if n is None:
n = int(width * height)
self.n = n
def reset(self):
pass
def sample(self):
return np.array([np.random.uniform(0, width, size=self.n),
np.random.uniform(0, height, size=self.n)]).T
# domain size, minimum distance between samples for Poisson disc method...
width = height = 100
r = 2
poisson_disc = PoissonDisc(width, height, r)
# Expected number of samples from Poisson disc method...
n = int(width * height / np.pi / poisson_disc.a**2)
# ... use the same for uniform noise.
uniform_noise = UniformNoise(width, height, n)
# Number of sampling runs to do (to remove noise from the noise in the power
# spectrum).
N = 100
# Sampling parameter, when putting the sample points onto the domain
M = 5
fig, ax = plt.subplots(nrows=2, ncols=2)
for j, noise in enumerate((poisson_disc, uniform_noise)):
print(noise.__class__.__name__)
spec = np.zeros((height * M, width * M))
for i in range(N):
print('{}/{}'.format(i+1, N))
noise.reset()
samples = np.array(noise.sample())
domain = np.zeros((height * M, width * M))
for pt in samples:
coords = int(pt[1] * M), int(pt[0] * M)
domain[coords] = 1
# Do the Fourier Trasform, shift the frequencies and add to the
# running total.
f = np.fft.fft2(domain)
fshift = np.fft.fftshift(f)
spec += np.log(np.abs(fshift))
# Plot the a set of random points and the power spectrum.
ax[0][j].imshow(domain, cmap=plt.cm.Greys)
ax[1][j].imshow(spec, cmap=plt.cm.Greys_r)
# Remove axis ticks and annotations
for k in (0,1):
ax[k][j].tick_params(which='both', bottom='off', left='off',
top='off', right='off', labelbottom='off', labelleft='off')
plt.savefig('periodograms.png')
The class PoissonDisc is defined in the file poisson.py given here:
import numpy as np
import matplotlib.pyplot as plt
class PoissonDisc():
def __init__(self, width=50, height=50, r=1, k=30):
self.width, self.height = width, height
self.r = r
self.k = k
# Cell side length
self.a = r/np.sqrt(2)
# Number of cells in the x- and y-directions of the grid
self.nx, self.ny = int(width / self.a) + 1, int(height / self.a) + 1
self.reset()
def reset(self):
"""Reset the cells dictionary."""
# A list of coordinates in the grid of cells
coords_list = [(ix, iy) for ix in range(self.nx)
for iy in range(self.ny)]
# Initilalize the dictionary of cells: each key is a cell's coordinates
# the corresponding value is the index of that cell's point's
# coordinates in the samples list (or None if the cell is empty).
self.cells = {coords: None for coords in coords_list}
def get_cell_coords(self, pt):
"""Get the coordinates of the cell that pt = (x,y) falls in."""
return int(pt[0] // self.a), int(pt[1] // self.a)
def get_neighbours(self, coords):
"""Return the indexes of points in cells neighbouring cell at coords.
For the cell at coords = (x,y), return the indexes of points in the
cells with neighbouring coordinates illustrated below: ie those cells
that could contain points closer than r.
ooo
ooooo
ooXoo
ooooo
ooo
"""
dxdy = [(-1,-2),(0,-2),(1,-2),(-2,-1),(-1,-1),(0,-1),(1,-1),(2,-1),
(-2,0),(-1,0),(1,0),(2,0),(-2,1),(-1,1),(0,1),(1,1),(2,1),
(-1,2),(0,2),(1,2),(0,0)]
neighbours = []
for dx, dy in dxdy:
neighbour_coords = coords[0] + dx, coords[1] + dy
if not (0 <= neighbour_coords[0] < self.nx and
0 <= neighbour_coords[1] < self.ny):
# We're off the grid: no neighbours here.
continue
neighbour_cell = self.cells[neighbour_coords]
if neighbour_cell is not None:
# This cell is occupied: store the index of the contained point
neighbours.append(neighbour_cell)
return neighbours
def point_valid(self, pt):
"""Is pt a valid point to emit as a sample?
It must be no closer than r from any other point: check the cells in
its immediate neighbourhood.
"""
cell_coords = self.get_cell_coords(pt)
for idx in self.get_neighbours(cell_coords):
nearby_pt = self.samples[idx]
# Squared distance between candidate point, pt, and this nearby_pt.
distance2 = (nearby_pt[0]-pt[0])**2 + (nearby_pt[1]-pt[1])**2
if distance2 < self.r**2:
# The points are too close, so pt is not a candidate.
return False
# All points tested: if we're here, pt is valid
return True
def get_point(self, refpt):
"""Try to find a candidate point near refpt to emit in the sample.
We draw up to k points from the annulus of inner radius r, outer radius
2r around the reference point, refpt. If none of them are suitable
(because they're too close to existing points in the sample), return
False. Otherwise, return the pt.
"""
i = 0
while i < self.k:
rho, theta = (np.random.uniform(self.r, 2*self.r),
np.random.uniform(0, 2*np.pi))
pt = refpt[0] + rho*np.cos(theta), refpt[1] + rho*np.sin(theta)
if not (0 <= pt[0] < self.width and 0 <= pt[1] < self.height):
# This point falls outside the domain, so try again.
continue
if self.point_valid(pt):
return pt
i += 1
# We failed to find a suitable point in the vicinity of refpt.
return False
def sample(self):
"""Poisson disc random sampling in 2D.
Draw random samples on the domain width x height such that no two
samples are closer than r apart. The parameter k determines the
maximum number of candidate points to be chosen around each reference
point before removing it from the "active" list.
"""
# Pick a random point to start with.
pt = (np.random.uniform(0, self.width),
np.random.uniform(0, self.height))
self.samples = [pt]
# Our first sample is indexed at 0 in the samples list...
self.cells[self.get_cell_coords(pt)] = 0
# and it is active, in the sense that we're going to look for more
# points in its neighbourhood.
active = [0]
# As long as there are points in the active list, keep looking for
# samples.
while active:
# choose a random "reference" point from the active list.
idx = np.random.choice(active)
refpt = self.samples[idx]
# Try to pick a new point relative to the reference point.
pt = self.get_point(refpt)
if pt:
# Point pt is valid: add it to samples list and mark as active
self.samples.append(pt)
nsamples = len(self.samples) - 1
active.append(nsamples)
self.cells[self.get_cell_coords(pt)] = nsamples
else:
# We had to give up looking for valid points near refpt, so
# remove it from the list of "active" points.
active.remove(idx)
return self.samples