Poisson disc sampling in Python
Posted on 22 April 2017
A set of points drawn from a uniform distribution on a two-dimensional domain typically display clustering:
import numpy as np
import matplotlib.pyplot as plt
width, height = 60, 45
N = width * height / 4
plt.scatter(np.random.uniform(0,width,N), np.random.uniform(0,height,N),
c='g', alpha=0.6, lw=0)
plt.xlim(0,width)
plt.ylim(0,height)
plt.axis('off')
plt.show()
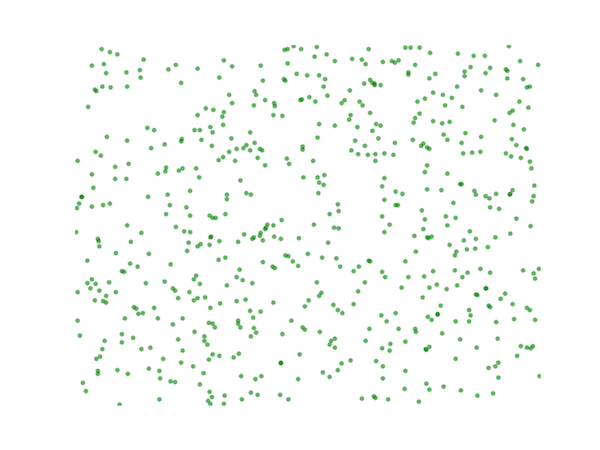
For computer graphics applications, it is often useful to obtain a sample of random points such that no one point is closer than some pre-determined distance from any other point, in order to avoid this clustering effect. This type of distribution, with no low-frequency components is sometimes called "blue noise".
A popular approach for obtaining non-clustered random sample of points is "poisson disc sampling"; an efficient ($O(n)$) algorithm to implement this approach was given by Bridson (ACM SIGGRAPH 2007 sketches, article 22)[pdf].
In a two-dimensional implementation of Bridson's algorithm, the sample $\boldsymbol{\mathrm{R}}^2$ domain is divided into square cells of side length $r/\sqrt{2}$ where $r$ is the minimum distance between samples, such that each cell can contain a maximum of one sample point. The sample points are stored in a list of $(x,y)$ coordinates, samples. The grid of cells are represented by a Python dictionary, cells, for which each key is the cell coordinates and the corresponding value is the index of the point in samples list (or None if the cell is empty).
We start by selecting an initial sample point (drawn at random uniformly from the domain), inserting it into samples and putting its index, 0, in the corresponding entry in the cells dictionary. We also initialize a separate list active with this index.
While the active list contains entries, we choose one at random, refpt, and generate up to $k$ (say, 30) points uniformly from the circular annulus around it of inner radius $r$ and outer radius $2r$. Any one of these points which is no closer than $r$ to any other in samples is "valid" and can be added to samples and active. We only need to check the surrounding cells in the local neighbourhood of refpt. If none of the $k$ points is valid, then refpt is removed from the active list: we will no longer search for points around this reference point.
Mike Bostock gives a nice animated demonstration of the Poisson disc sampling algorithm on his website.
Below is my Python code for Poisson disc sampling using Bridson's algorithm; a typical output is shown here:
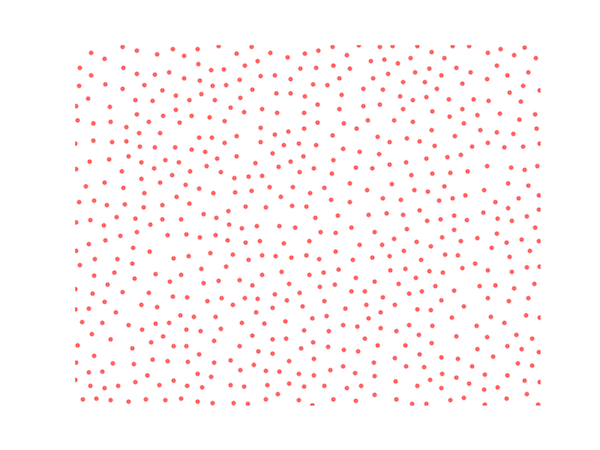
Please see the next post for an object-oriented approach to this algorithm.
This code is also available on my github page.
import numpy as np
import matplotlib.pyplot as plt
# Choose up to k points around each reference point as candidates for a new
# sample point
k = 30
# Minimum distance between samples
r = 1.7
width, height = 60, 45
# Cell side length
a = r/np.sqrt(2)
# Number of cells in the x- and y-directions of the grid
nx, ny = int(width / a) + 1, int(height / a) + 1
# A list of coordinates in the grid of cells
coords_list = [(ix, iy) for ix in range(nx) for iy in range(ny)]
# Initilalize the dictionary of cells: each key is a cell's coordinates, the
# corresponding value is the index of that cell's point's coordinates in the
# samples list (or None if the cell is empty).
cells = {coords: None for coords in coords_list}
def get_cell_coords(pt):
"""Get the coordinates of the cell that pt = (x,y) falls in."""
return int(pt[0] // a), int(pt[1] // a)
def get_neighbours(coords):
"""Return the indexes of points in cells neighbouring cell at coords.
For the cell at coords = (x,y), return the indexes of points in the cells
with neighbouring coordinates illustrated below: ie those cells that could
contain points closer than r.
ooo
ooooo
ooXoo
ooooo
ooo
"""
dxdy = [(-1,-2),(0,-2),(1,-2),(-2,-1),(-1,-1),(0,-1),(1,-1),(2,-1),
(-2,0),(-1,0),(1,0),(2,0),(-2,1),(-1,1),(0,1),(1,1),(2,1),
(-1,2),(0,2),(1,2),(0,0)]
neighbours = []
for dx, dy in dxdy:
neighbour_coords = coords[0] + dx, coords[1] + dy
if not (0 <= neighbour_coords[0] < nx and
0 <= neighbour_coords[1] < ny):
# We're off the grid: no neighbours here.
continue
neighbour_cell = cells[neighbour_coords]
if neighbour_cell is not None:
# This cell is occupied: store this index of the contained point.
neighbours.append(neighbour_cell)
return neighbours
def point_valid(pt):
"""Is pt a valid point to emit as a sample?
It must be no closer than r from any other point: check the cells in its
immediate neighbourhood.
"""
cell_coords = get_cell_coords(pt)
for idx in get_neighbours(cell_coords):
nearby_pt = samples[idx]
# Squared distance between or candidate point, pt, and this nearby_pt.
distance2 = (nearby_pt[0]-pt[0])**2 + (nearby_pt[1]-pt[1])**2
if distance2 < r**2:
# The points are too close, so pt is not a candidate.
return False
# All points tested: if we're here, pt is valid
return True
def get_point(k, refpt):
"""Try to find a candidate point relative to refpt to emit in the sample.
We draw up to k points from the annulus of inner radius r, outer radius 2r
around the reference point, refpt. If none of them are suitable (because
they're too close to existing points in the sample), return False.
Otherwise, return the pt.
"""
i = 0
while i < k:
i += 1
rho = np.sqrt(np.random.uniform(r**2, 4 * r**2))
theta = np.random.uniform(0, 2*np.pi)
pt = refpt[0] + rho*np.cos(theta), refpt[1] + rho*np.sin(theta)
if not (0 <= pt[0] < width and 0 <= pt[1] < height):
# This point falls outside the domain, so try again.
continue
if point_valid(pt):
return pt
# We failed to find a suitable point in the vicinity of refpt.
return False
# Pick a random point to start with.
pt = (np.random.uniform(0, width), np.random.uniform(0, height))
samples = [pt]
# Our first sample is indexed at 0 in the samples list...
cells[get_cell_coords(pt)] = 0
# ... and it is active, in the sense that we're going to look for more points
# in its neighbourhood.
active = [0]
nsamples = 1
# As long as there are points in the active list, keep trying to find samples.
while active:
# choose a random "reference" point from the active list.
idx = np.random.choice(active)
refpt = samples[idx]
# Try to pick a new point relative to the reference point.
pt = get_point(k, refpt)
if pt:
# Point pt is valid: add it to the samples list and mark it as active
samples.append(pt)
nsamples += 1
active.append(len(samples)-1)
cells[get_cell_coords(pt)] = len(samples) - 1
else:
# We had to give up looking for valid points near refpt, so remove it
# from the list of "active" points.
active.remove(idx)
plt.scatter(*zip(*samples), color='r', alpha=0.6, lw=0)
plt.xlim(0, width)
plt.ylim(0, height)
plt.axis('off')
plt.show()