Isolines on a nuclide chart
Posted on 26 April 2021
As a short addendum to this blog post, the code below plots a chart of the nuclides showing four types of isoline:
- isotopes: these are nuclides with the same proton number, $Z$ (and hence nuclear charge) – vertical lines in the chart.
- isotones: nuclides with the same number of neutrons, $N$ – horizontal lines.
- isobars: nuclides with the same mass number, $N+Z$: beta decay converts between isobaric nuclides by transforming a neutron into a proton ($\beta^-$ decay) or a proton into a neutron ($\beta^+$ decay) – these are the blue diagonal lines.
- isodiaphers: nuclides with the same neutron excess, $N-Z$: since an alpha particle consists of two neutrons and two protons, when a nuclide undergoes alpha decay, the product nucleus is an isodiapher of the original – magenta diagonal lines.
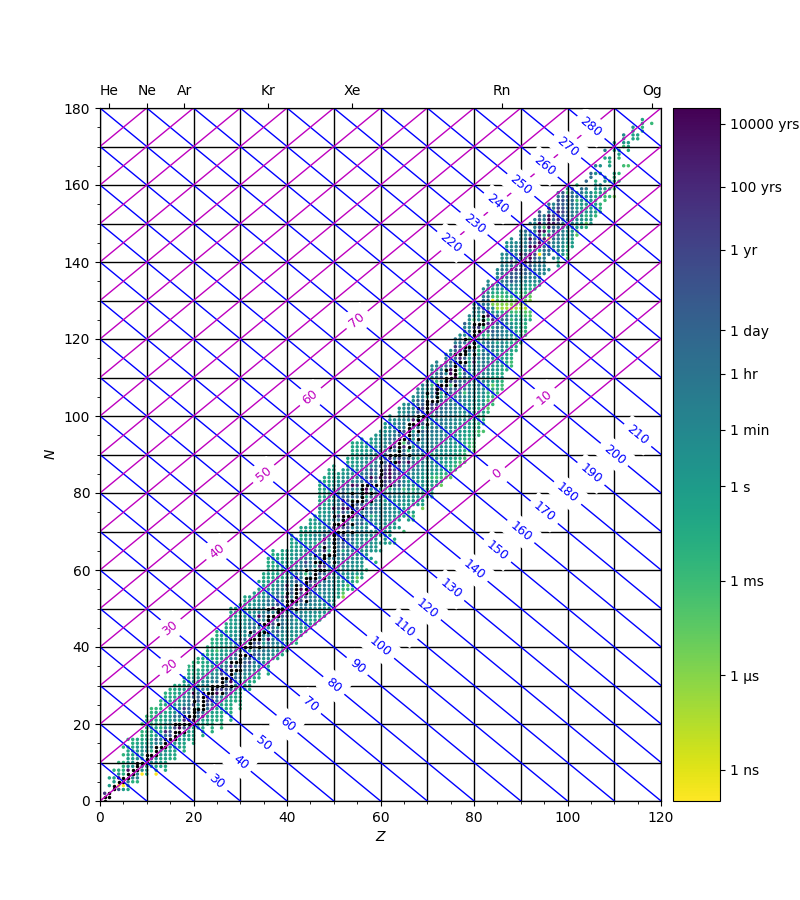
The code below requires the file halflives.txt.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import Normalize
from matplotlib.cm import ScalarMappable
from matplotlib import ticker
PTS_PER_INCH = 72
element_symbols = [
'H', 'He', 'Li', 'Be', 'B', 'C', 'N', 'O', 'F', 'Ne', 'Na', 'Mg', 'Al',
'Si', 'P', 'S', 'Cl', 'Ar', 'K', 'Ca', 'Sc', 'Ti', 'V', 'Cr', 'Mn', 'Fe',
'Co', 'Ni', 'Cu', 'Zn', 'Ga', 'Ge', 'As', 'Se', 'Br', 'Kr', 'Rb', 'Sr', 'Y',
'Zr', 'Nb', 'Mo', 'Tc', 'Ru', 'Rh', 'Pd', 'Ag', 'Cd', 'In', 'Sn', 'Sb', 'Te',
'I', 'Xe', 'Cs', 'Ba', 'La', 'Ce', 'Pr', 'Nd', 'Pm', 'Sm', 'Eu', 'Gd', 'Tb',
'Dy', 'Ho', 'Er', 'Tm', 'Yb', 'Lu', 'Hf', 'Ta', 'W', 'Re', 'Os', 'Ir', 'Pt',
'Au', 'Hg', 'Tl', 'Pb', 'Bi', 'Po', 'At', 'Rn', 'Fr', 'Ra', 'Ac', 'Th', 'Pa',
'U', 'Np', 'Pu', 'Am', 'Cm', 'Bk', 'Cf', 'Es', 'Fm', 'Md', 'No', 'Lr', 'Rf',
'Db', 'Sg', 'Bh', 'Hs', 'Mt', 'Ds', 'Rg', 'Cn', 'Nh', 'Fl', 'Mc', 'Lv', 'Ts',
'Og'
]
def get_marker_size(ax, nx, ny):
"""Determine the appropriate marker size (in points-squared) for ax."""
# Get Axes width and height in pixels.
bbox = ax.get_window_extent().transformed(fig.dpi_scale_trans.inverted())
width, height = bbox.width, bbox.height
# Spacing between scatter point centres along the narrowest dimension.
spacing = min(width, height) * PTS_PER_INCH / min(nx, ny)
# Desired radius of scatter points.
rref = spacing / 2 * 0.5
# Initial scatter point size (area) in pt^2.
s = np.pi * rref**2
return s
def annotate_line(x, y, label, xylabel=None, colour='k'):
"""Annotate an isobar or isodiapher line.
The text label is rotated to coincide with the line through
(x[0], y[0]) and (x[1], y[1]) and centred at xylabel. If not provided,
xylabel is set to the centre of the line.
"""
rotn = np.degrees(np.arctan2(y[1]-y[0], x[1]-x[0]))
if xylabel is None:
xylabel = ((x[0]+x[1])/2, (y[0]+y[1])/2)
text = ax.annotate(label, xy=xylabel, ha='center', va='center',
backgroundcolor='white', color=colour, size=9)
p1 = ax.transData.transform_point((x[0], y[0]))
p2 = ax.transData.transform_point((x[1], y[1]))
dy = (p2[1] - p1[1])
dx = (p2[0] - p1[0])
rotn = np.degrees(np.arctan2(dy, dx))
text.set_rotation(rotn)
# Read in the data, determining the nuclide half-lives as a function of the
# number of neutrons, N, and protons, Z. Stable nuclides are indicated with
# -1 in the half-life field: deal with these separately from the radioactive
# nuclei.
halflives = {}
stables = []
for line in open('halflives.txt'):
line = line.rstrip()
halflife = line[10:]
if halflife == 'None':
continue
fields = line[:10].split('-')
symbol = fields[0]
A = int(fields[1].split()[0])
Z = int(element_symbols.index(symbol)) + 1
N = A - Z
halflife = float(halflife)
if halflife < 0:
stables.append((N, Z))
else:
halflives[(N, Z)] = halflife
# Unwrap the halflives dictionary into sequences of N, Z, thalf; take the log.
k, thalf = zip(*halflives.items())
N, Z = zip(*k)
maxN, maxZ = 180, 120
log_thalf = np.log10(thalf)
# Initial dimensions of the plot (pixels); figure resolution (dots-per-inch).
w, h = 800, 900
DPI = 100
w_in, h_in = w / DPI, h / DPI
# Set up the figure: one Axes object for the plot, another for the colourbar.
fig, (ax, cax) = plt.subplots(1, 2,
gridspec_kw={'width_ratios': [12, 1], 'wspace': 0.04},
figsize=(w_in, h_in), dpi=DPI)
# Pick a colourmap and set the normalization: nuclides with half-lives of less
# than 10^vmin or greater than 10^vmax secs are off-scale.
cmap = plt.get_cmap('viridis_r')
norm = Normalize(vmin=-10, vmax=12)
colours = cmap(norm(log_thalf))
s = get_marker_size(ax, maxZ, maxN)
sc1 = ax.scatter(Z, N, c=colours, s=s)
# Stable nuclides are plotted in black ('k').
Nstable, Zstable = zip(*stables)
sc2 = ax.scatter(Zstable, Nstable, c='k', s=s, label='stable')
loc = ticker.MultipleLocator(base=5)
ax.xaxis.set_minor_locator(loc)
ax.yaxis.set_minor_locator(loc)
Ngrid = np.linspace(0, maxN, 19)
# Isotopes: lines of constant Z.
ax.vlines(np.linspace(0, maxZ, 13), 0, maxN, lw=1, color='k')
# Isotones: lines of constant N.
ax.hlines(Ngrid, 0, maxZ, lw=1, color='k')
# Isobars: lines of constant mass number, N + Z.
isobar_grid = np.linspace(0, maxN + maxZ, 31, dtype=int)
for NplusZ in isobar_grid:
x, y = [0, NplusZ], [NplusZ, 0]
ax.plot(x, y, lw=1, color='b')
if 30 <= NplusZ < 290:
if NplusZ <= 210:
xlabel = 0.5 * NplusZ + 10
else:
xlabel = 0.5 * NplusZ - 35
ylabel = NplusZ - xlabel
label = str(NplusZ)
xylabel = (xlabel, ylabel)
annotate_line(x, y, label, xylabel=xylabel, colour='b')
# Isodiaphers: lines of constant neutron excess, N - Z.
isodiapher_grid = np.linspace(0, maxN, 19, dtype=int)
# Hand-picked positions for the isodiapher labels
xlabel = [85, 95, 15, 15, 25, 35, 45, 55]
for i, NminusZ in enumerate(isodiapher_grid):
x, y = [0, maxZ], [NminusZ, maxZ + NminusZ]
ax.plot(x, y, lw=1, color='m')#color=(0.5, 0.5, 0.5))
if i < len(xlabel):
ylabel = xlabel[i] + NminusZ
label = str(NminusZ)
annotate_line(x, y, label, xylabel=(xlabel[i], ylabel), colour='m')
ax.set_xlim(0, maxZ)
ax.set_ylim(0, maxN)
ax.set_xlabel(r'$Z$')
ax.set_ylabel(r'$N$')
# Twin the y-axis to indicate the noble gas elements on the top x-axis.
topax = ax.twiny()
topax.set_xticks([2, 10, 18, 36, 54, 86, 118])
topax.set_xticklabels(['He', 'Ne', 'Ar', 'Kr', 'Xe', 'Rn', 'Og'])
topax.set_xlim(0, maxZ)
# The colourbar: label key times in appropriate units.
cbar = fig.colorbar(ScalarMappable(norm=norm, cmap=cmap), cax=cax)
SECS_PER_DAY = 24 * 3600
SECS_PER_YEAR = 365.2425 * SECS_PER_DAY
cbar_ticks = np.log10([1.e-9, 1.e-6, 1.e-3, 1, 60, 3600, SECS_PER_DAY,
SECS_PER_YEAR, 100 * SECS_PER_YEAR, 10000 * SECS_PER_YEAR])
cbar_ticklabels = ['1 ns', '1 μs', '1 ms', '1 s', '1 min', '1 hr', '1 day',
'1 yr', '100 yrs', '10000 yrs']
cbar.set_ticks(cbar_ticks)
cbar.set_ticklabels(cbar_ticklabels)
def on_resize(event):
"""Callback function on interactive plot window resize."""
# Resize the scatter plot markers to keep the plot looking good.
s = get_marker_size(ax, maxZ, maxN)
sc1.set_sizes([s]*len(Z))
sc2.set_sizes([s]*len(stables))
# Attach the callback function to the window resize event.
cid = fig.canvas.mpl_connect('resize_event', on_resize)
plt.savefig('nuclide-isolines.png', dpi=DPI)
plt.show()