The two-dimensional wave equation
Posted on 17 February 2024
The wave equation is a second-order linear partial differential equation describing the behaviour of mechanical waves; its two (spatial) dimensional form can be used to describe waves on a surface of water:
$$ \frac{\partial^2 u}{\partial t^2} - c^2\left( \frac{\partial^2 u}{\partial x^2} + \frac{\partial^2 u}{\partial y^2} \right) = 0 $$
To model such waves numerically, it is common to work with a discrete grid of spatial and time points and to approximate the partial derivatives using the method of finite differences. A simple approach is to take the central difference using neighbouring points on the grid. In one dimension, for a grid spacing of $h$, the first and second derivatives are approximately:
$$ \frac{\mathrm{d}f}{\mathrm{d}x} \approx \frac{f(x + \frac{h}{2}) - f(x - \frac{h}{2})}{h} $$
and $$ \begin{align*} \frac{\mathrm{d}^2f}{\mathrm{d}x^2} & \approx \frac{\frac{f(x + h) - f(x)}{h} - \frac{f(x) - f(x - h)}{h}}{h}\\ &= \frac{f(x+h) - 2f(x) + f(x-h)}{h^2}. \end{align*} $$ In two dimensions (assuming equal grid spacing in each direction), the most basic numerical approach is to use the five-point stencil: which amounts to the following: $$ \begin{align*} \frac{\partial^2 f}{\partial x^2} \approx \frac{f(x+h, y) - 2f(x, y) + f(x-h, y)}{h^2}\\ \frac{\partial^2 f}{\partial y^2} \approx \frac{f(x, y+h) - 2f(x, y) + f(x, y-h)}{h^2}\\ \end{align*} $$ Therefore, the procedure is to choose a spatial grid size, $h$, and time step size, $\delta_t$, and represent the function as: $u(t; x, y) = u_{i,j}^{(k)}$ where $k$ labels the time step: $u(t+\delta_t; x, y) = u_{i,j}^{(k+1)}$ and $i$ and $j$ the $x$ and $y$ coordinates of the spatial grid: e.g., $u(t, x+h, y) = u_{i+1, j}^{(k)}$, $u(t, x, y+h) = u_{i, j+1}^{(k)}$ etc.
The above finite difference equations then approximate the wave equation as:
$$ \frac{1}{\delta_t^2}\left( u_{i,j}^{(k+1)} - 2 u_{i,j}^{(k)} + u_{i,j}^{(k-1)} \right) - \frac{c^2}{h^2} \left( u_{i+1,j}^{(k)} + u_{i-1,j}^{(k)} + u_{i,j+1}^{(k)} + u_{i,j-1}^{(k)} - 4 u_{i,j}^{(k)} \right) = 0 $$
The goal of the simulation is to predict how the wave function $u$ will evolve in time for each point on the spatial grid: i.e. to find $u_{i,j}^{(k+1)}$:
$$ u_{i,j}^{(k+1)} = \alpha^2\left( u_{i+1,j}^{(k)} + u_{i-1,j}^{(k)} + u_{i,j+1}^{(k)} + u_{i,j-1}^{(k)} - 4 u_{i,j}^{(k)} \right) + 2u_{i,j}^{(k)} - u_{i,j}^{(k-1)}, $$
where $\alpha = c\delta_t/h$ .This would be the end of the story if the spatial grid had infinite extent, but in practice we have to choose a finite number of $(x,y)$ points and therefore need to worry about what happens at the boundary of the grid. One choice is simply to fix the boundary values of the wave function to be zero: $u_{0,j} = u_{i, 0} = u_{N_x, j} = u_{i, N_y} = 0$. This is a Dirichlet boundary condition and means that no wave energy leaves the simulation grid: the waves are reflected back.
An alternative choice is an absorbing boundary condition (ABC), for which no reflection occurs: there are different ways of approximating this condition, but a popular one is the Mur boundary condition. This can be demonstrated in one-dimension by factorizing the wave equation as
$$ \frac{\partial^2 u}{\partial t^2} - c^2\frac{\partial^2 u}{\partial x^2} = \left( \frac{\partial}{\partial t} - c\frac{\partial}{\partial x}\right) \left( \frac{\partial}{\partial t} + c\frac{\partial}{\partial x}\right)u = 0, $$
Each factor represents a "one-way" wave equation since they correspond to equations with solutions traveling in the $-x$ and $+x$ directions: $$ \begin{align*} \frac{\partial u}{\partial t} - c\frac{\partial u}{\partial x} = 0 \Rightarrow & \; u_\leftarrow = e^{i(kx+\omega t)},\\ \frac{\partial u}{\partial t} + c\frac{\partial u}{\partial x} = 0 \Rightarrow & \; u_\rightarrow = e^{i(kx-\omega t)}. \end{align*} $$ We therefore apply the first of these equations at the $x=0$ boundary so that only waves traveling out of the domain in the negative $x$ direction are supported (no reflection back into the domain); at the other boundary, we apply the second equation to ensure that the solution consists solely of waves travelling out of the domain in the positive $x$ direction.
In the discretized versions of these equations there is a complication in that the spatial and time derivatives have to be evaluated at the same point (in time and space), e.g. for the left hand boundary:
$$ u_{\frac{1}{2},j}^{(k+1)} - u_{\frac{1}{2},j}^{(k)} - \frac{c\delta_t}{h} \left( u_{1,j}^{\left(k+\frac{1}{2}\right)} - u_{0,j}^{\left(k+\frac{1}{2}\right)} \right) = 0. $$
Of course, we don't have half-integer indexes for our space and time steps, so instead choose to take the average of the neighbouring points:
$$ \frac{1}{2}\left( \frac{u_{1,j}^{(k+1)} - u_{1,j}^{(k)}}{\delta_t} + \frac{u_{0,j}^{(k+1)} - u_{0,j}^{(k)}}{\delta_t}\right) - \frac{c}{2} \left( \frac{u_{1,j}^{(k+1)} - u_{0,j}^{(k+1)}}{h} + \frac{u_{1,j}^{(k)} - u_{0,j}^{(k)}}{h} \right) = 0 $$
Rearranging for the required quantity, $u_{0,j}^{(k+1)}$ gives:
$$ u_{0,j}^{(k+1)} = u_{1,j}^{(k)} - \frac{1-\alpha}{1+\alpha} \left( u_{1,j}^{(k+1)} - u_{0,j}^{(k)}\right). $$
Similarly, at the right hand boundary:
$$ u_{N,j}^{(k+1)} = u_{N-1,j}^{(k)} + \frac{1-\alpha}{1+\alpha}\left( u_{N,j}^{(k)} - u_{N-1,j}^{(k+1)} \right), $$
where $N$ is the index of the final coordinate in the $x$ direction (here, $N=n_x-1$ because of Python's zero-based indexing). Corresponding equations apply for the top and bottom boundaries in the $y$ direction.
The class below implements this integration scheme for the two-dimensional wave equation.
import numpy as np
class WaveEqn2D:
def __init__(self, nx=500, ny=500, c=0.2, h=1, dt=1,
use_mur_abc=True):
"""Initialize the simulation:
nx and ny are the dimension of the domain;
c is the wave speed;
h and dt are the space and time grid spacings;
If use_mur_abc is True, the Mur absorbing boundary
conditions will be used; if False, the Dirichlet
(reflecting) boundary conditions are used.
"""
self.nx, self.ny = nx, ny
self.c = c
self.h, self.dt = 1, 1
self.use_mur_abc = use_mur_abc
self.alpha = self.c * self.dt / self.h
self.alpha2 = self.alpha**2
self.u = np.zeros((3, ny, nx))
def update(self):
"""Update the simulation by one time tick."""
# The three planes of u correspond to the time points
# k+1, k and k-1; i.e. we calculate the next frame
# of the simulation (k+1) in u[0,...].
u, nx, ny = self.u, self.nx, self.ny
u[2] = u[1] # old k -> new k-1
u[1] = u[0] # old k+1 -> new k
# Calculate the new k+1:
u[0, 1:ny-1, 1:nx-1] = self.alpha2 * (
u[1, 0:ny-2, 1:nx-1]
+ u[1, 2:ny, 1:nx-1]
+ u[1, 1:ny-1, 0:nx-2]
+ u[1, 1:ny-1, 2:nx]
- 4*u[1, 1:ny-1, 1:nx-1]) \
+ (2 * u[1, 1:ny-1, 1:nx-1]
- u[2, 1:ny-1, 1:nx-1])
if self.use_mur_abc:
# Mur absorbing boundary conditions.
kappa = (1 - self.alpha) / (1 + self.alpha)
u[0, 0, 1:nx-1] = (u[1, 1, 1:nx-1]
- kappa * (
u[0, 1, 1:nx-1]
- u[1, 0, 1:nx-1])
)
u[0, ny-1, 1:nx-1] = (u[1, ny-2, 1:nx-1]
+ kappa * (
u[1, ny-1, 1:nx-1]
- u[0, ny-2, 1:nx-1])
)
u[0, 1:ny-1, 0] = (u[1, 1:ny-1, 1]
- kappa * (
u[0, 1:ny-1, 1]
- u[1, 1:ny-1, 0])
)
u[0, 1:ny-1, nx-1] = (u[1, 1:ny-1, nx-2]
+ kappa * (
u[1, 1:ny-1, nx-1]
- u[0, 1:ny-1, nx-2])
)
Its use is illustrated below for modelling the waves produced by a signal, sinusoidally varying in time at the centre of the domain.
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
from wave_eqn2d import WaveEqn2D
A = 80
dt = 1
T = 50
freq = 2 * np.pi / T
nx = ny = 200
sim = WaveEqn2D(nx, ny, dt=dt, use_mur_abc=True)
fig, ax = plt.subplots()
ax.axis("off")
img = ax.imshow(sim.u[0], vmin=0, vmax=40, cmap='Blues_r')
def update(i):
"""Advance the simulation by one tick."""
# A regular sinusoidal signal at the centre of the domain.
sim.u[0, ny//2, nx//2] = A * np.sin(i * freq)
sim.update()
def init():
"""
Initialization, because we're blitting and need references to the
animated objects.
"""
return img,
def animate(i):
"""Draw frame i of the animation."""
update(i)
img.set_data(sim.u[0])
return img,
interval, nframes = sim.dt, 1000
ani = animation.FuncAnimation(fig, animate, frames=nframes,
repeat=False,
init_func=init, interval=interval, blit=True)
plt.show()
The difference between the reflecting (Dirichlet) and absorbing (Mur) boundary conditions can be seen in the animations produced using use_mur_abc=False and use_mur_abc=True:
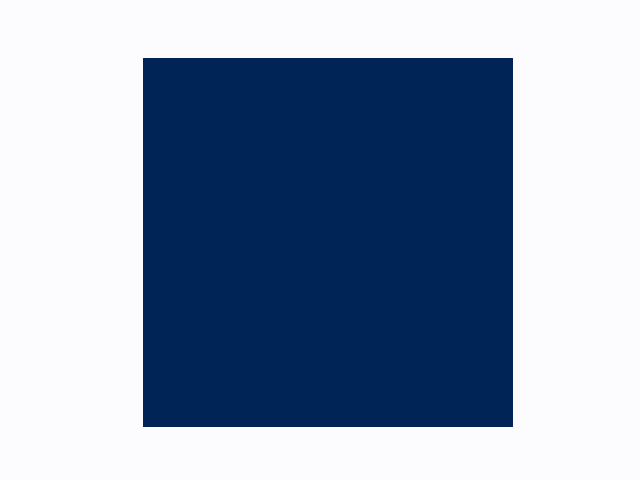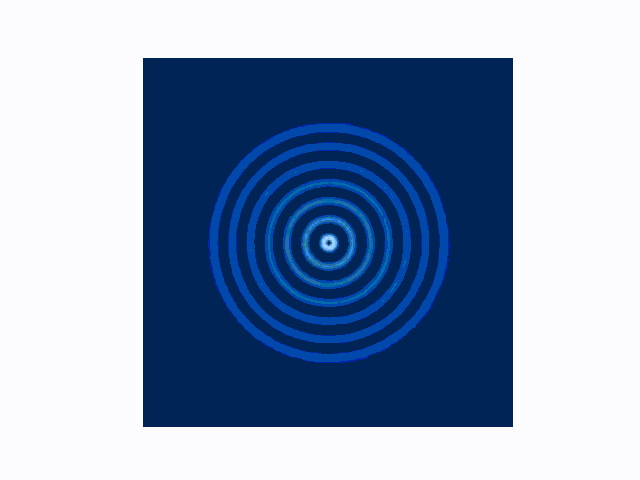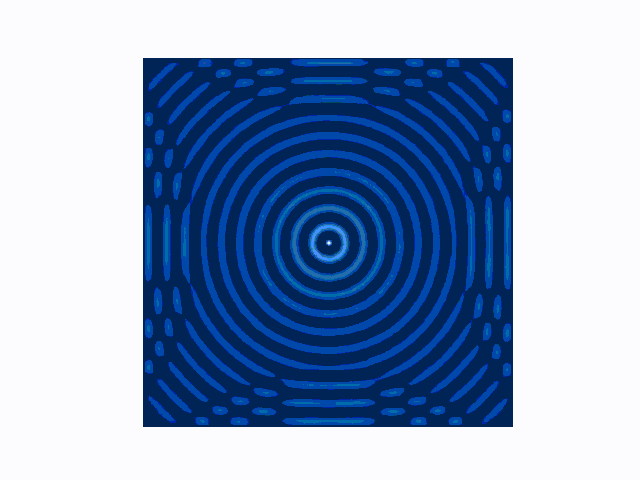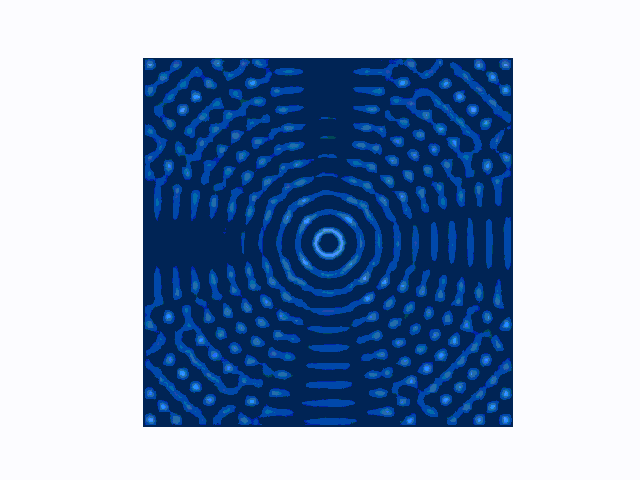
Dirichlet boundary conditions
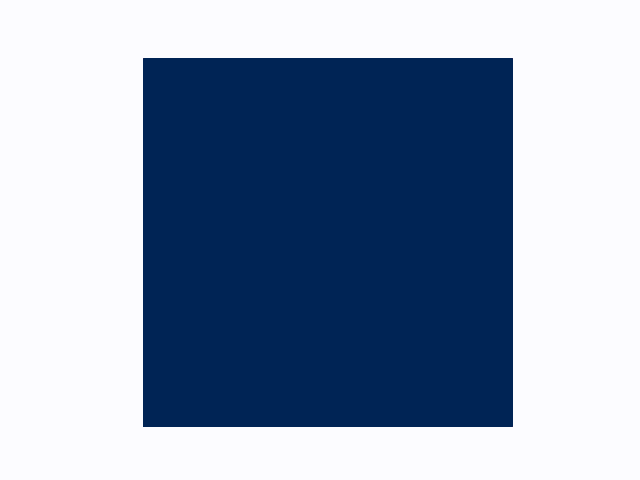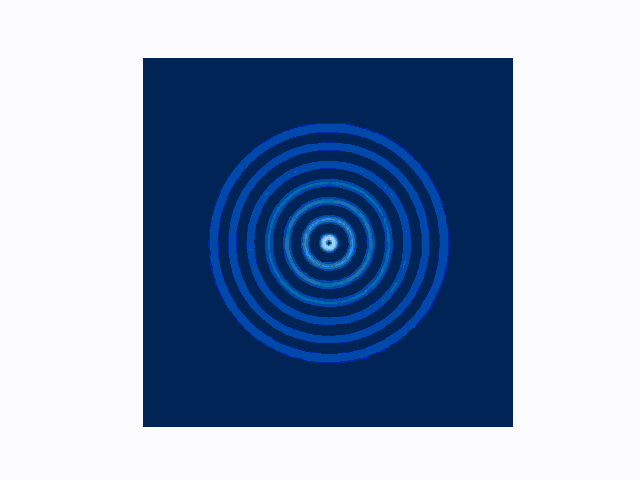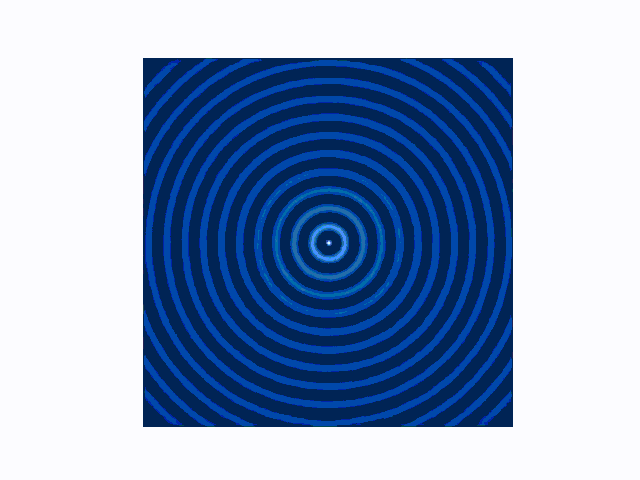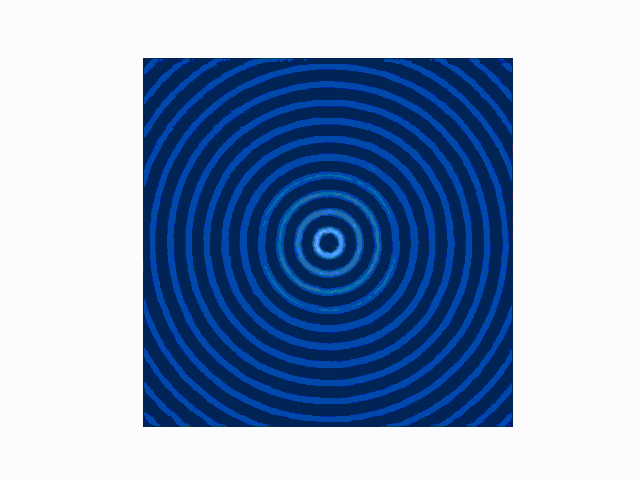
Mur absorbing boundary conditions
Another example, creating a raindrop effect by randomly placing two-dimensional Gaussian profiles on the domain:
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.animation as animation
from wave_eqn2d import WaveEqn2D
# Raindrop probability (with each time tick) and intensity.
drop_probability, max_intensity = 0.01, 10
# Width of the Gaussian profile for each initial drop.
drop_width = 2
# Number of Gaussian widths to calculate to.
ndrop_widths = 3
# Size of the Gaussian template each drop is based on.
NDx = NDy = drop_width * ndrop_widths
Dx, Dy = np.arange(0, NDx, 1, dtype=np.int32), np.arange(0, NDy, 1, dtype=np.int32)
MDx, MDy = np.meshgrid(Dx, Dy)
# Create the 2D template of the initial drop.
cx, cy = NDx // 2, NDy // 2
gauss_template = np.exp(-(((MDx-cx)/drop_width)**2 + ((MDy-cy)/drop_width)**2))
dt = 1
nx = ny = 200
sim = WaveEqn2D(nx, ny, dt=dt, use_mur_abc=True)
fig, ax = plt.subplots()
ax.axis("off")
img = ax.imshow(sim.u[0], vmin=0, vmax=max_intensity, cmap='YlGnBu_r')
def update(i):
"""Advance the simulation by one tick."""
# Random raindrops.
if np.random.random() < drop_probability:
x, y = np.random.randint(NDx//2, nx-NDx//2-1), np.random.randint(NDy//2, ny-NDy//2-1)
sim.u[0, y-NDy//2:y+NDy//2, x-NDx//2:x+NDx//2] = max_intensity * gauss_template
sim.update()
def init():
"""
Initialization, because we're blitting and need references to the
animated objects.
"""
return img,
def animate(i):
"""Draw frame i of the animation."""
update(i)
img.set_data(sim.u[0])
return img,
interval, nframes = 2*sim.dt, 4000
ani = animation.FuncAnimation(fig, animate, frames=nframes,
repeat=False,
init_func=init, interval=interval, blit=True)
plt.show()
